Welcome to cottonENCODE database
The primary objective of Encyclopedia of DNA Elements project (ENCODE) was to identify all functional element regions in the entire genome. The ultimate goal of this endeavor is to construct a comprehensive and dynamic multidimensional regulatory map representing multiple processes and tissues. Here, we initiated the inaugural cottonENCODE database, which involved the compilation of previously undocumented ultra-high resolution (3 Kb) in situ Hi-C chromatin contact maps, low background noise datasets of ChIP-Seq for two active histone modifications (H3K4me3 and H3K27ac), ATAC-Seq and RNA-Seq libraries from 12 different tissues and developmental stages of Gossypium hirsutum.
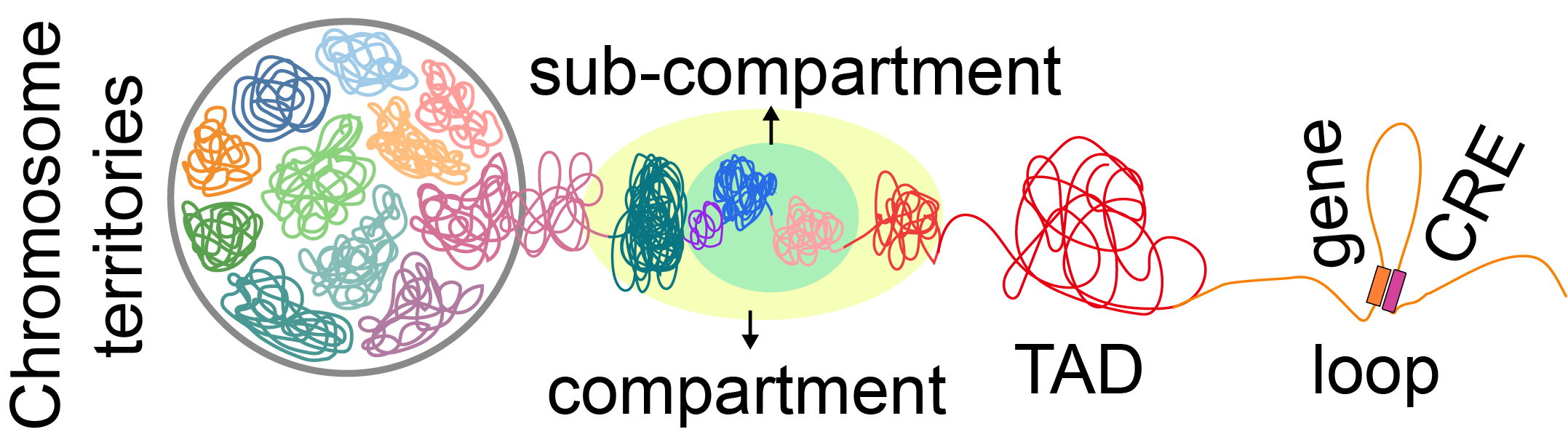
The database offers visualization functions for multi-omics data, enabling the comparison of various 3D genome feature structures (A/B compartment, sub-compartments, TAD-like domains and loop), histone modification sites (H3K4me3 and H3K27ac), open chromatin regions, and gene expression levels between different tissues. Moreover, users can access 3D feature structures and chromatin modification status of specific regions and genes by the search function. All datasets from this database are available for free download. In summary, the cottonENCODE database provides abundant resources and unlocks the potential of genomes, thereby facilitating the understanding for biological processes and guiding cotton breeding.